Please the eye and plague the heart
贪图-时快活, 必然留下隐患
Updated time: 06/24 2024.
Find the heterostructure
Tools
see Tool - Summary
Some talks / advices
ASE/Pymatgen
- stack/interface function - only used for some similar structure (lattice type, constant, orientation), but could found a minimal force on the interface with specified interlayer space
VASPKIT
When I use VASPKIT to generate a heterostructure, I found some interesting thing.
- VASPKIT can’t fix the lattice constant of upper or lower material.
- I found over-distorted material in some model of extremely small angle, causing lose its original lattice information like HCP/FCC/… I think it’s because the VASPKIT didn’t set a tolerance of angle mismatch, and it has important influence in this extremely small angle model.
VASPKIT First-Cup
Because I want to fix the lattice constant of substrates, the VASPKIT couldn’t meet this requirement.
The code of The First Prize - LUOJUN maybe could do it.
Dependency
- VASPKIT = 0.73
- Python3
- some python packages
Error
- VASPKIT must ~0.73, because the bash command has run
VASPKIT 406. This command is406 Converte POSCAR to other format -> POSCAR -> 4061 POSCAR with Cartesian Coordinatesin 0.73 version, but has been changed now.
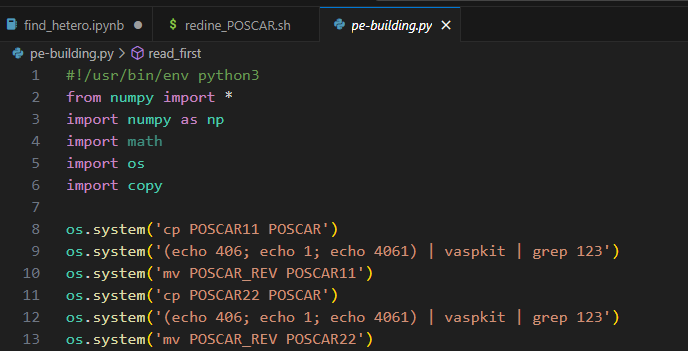
- The POSCAR can’t has
0.00000000direct coordinate, if it has, this procedure will output some unexpected value errors.

Parameters
- v is controlling the shifting basic vector, maybe could be using to search more heterostructures.
Hetbuilder
It has been installed !!!!
Installation
% python3 3.11.2
python3 -m venv /../../env
% activate
source /../../env/bin/activate
% get root
sudo apt-get update && sudo apt install -y build-essential cmake git python3-dev python3-tk libsymspg-dev
git clone https://github.com/hongyi-zhao/hetbuilder.git && cd hetbuilder
pip install -r requirements.txt
pip install .
The Coincidence lattices theory maybe too strict for high mismatched film/substrate system to search a minimal configuration (like vaspkit, ~45 atoms).
The most output has > 200 atoms, although we enlarge the t parameter - space in Angstrom unit.
usage (see more mymetal/build/findhetero.py)
hetbuilder --help
hetbuilder build --help
hetbuilder -N 2 -t 2 -w 0 CONTCAR_SiO_film CONTCAR_FCC_film
from hetbuilder.algorithm import CoincidenceAlgorithm
from hetbuilder.plotting import InteractivePlot
from ase.io.vasp import read_vasp, write_vasp
bottom = read_vasp('CONTCAR_SiO_film')
top = read_vasp('CONTCAR_FCC_film')
# we set up the algorithm class
alg = CoincidenceAlgorithm(bottom, top)
# we run the algorithm for a choice of parameters
results = alg.run(Nmax = 5, Nmin = 0,
tolerance = 2, weight = 0.5)
#for j in results:
# print(j)
#%matplotlib widget
import matplotlib
matplotlib.use('Qt5Agg') # if failed, pip install PyQt5 PySide2
iplot = InteractivePlot(bottom=bottom, top=top, results=results, weight=0)
iplot.plot_results()
j = results[1]
print(type(j))
print(j.bottom)
print(j.top)
print(j.stack)
print(j.strain)
# view(bottom)
# view(j.stack)
construct heterostructure
from ase import Atoms
from ase.build import make_supercell
from ase.build.tools import sort
from numpy import array, zeros, concatenate
from ase.visualize import view
def build_supercells(primitive_bottom: Atoms = None,
primitive_top: Atoms = None,
M: array = None,
N: array = None,
angle_z: float = None,
weight: float = 0.5,
distance: float = 3.5,
vacuum: float = 15,
pbc: list = [True, True, False],
reorder=True ) -> Atoms:
"""
For construct supercell for two primitive cell known transition matrix and rotation angle of top layer around z-dir\n
Input: must be primitive cell\n
M, N: bottom, top matrix\n
angle_z: units: degree, +z direction\n
distance, vacuum: units: Angstron\n
weight: must be 0~1, fixing bottom~top\n
pbc, reorder: the default value is good\n
Output:
heterostructure
Usage:
For output of hetbuilder, results is a list contained many structures.
defined: j = results[0] or any index
build_supercells(j.bottom, j.top, j.M, j.N, j.angle, j._weight) # note: _weight
For general situation,
build_supercells(bottom, top, M, N, angle, weight)
"""
bottom = primitive_bottom.copy()
top = primitive_top.copy()
#print(top)
# make_supercell
bottom_sup = make_supercell(bottom, M)
top_sup = make_supercell(top, N)
#print(bottom_sup)
# rotate top layer around z, units: degree
top_sup.rotate(angle_z, 'z', rotate_cell=True)
#print(top_sup)
# translate from hetbuilder (atom_functions.cpp/.h) stack_atoms function
stack = stack_atoms(bottom_sup, top_sup, weight, distance, vacuum, pbc, reorder)
#print(1)
return stack
def stack_atoms(bottom_sup: Atoms = None, top_sup: Atoms = None, weight: float = 0.5, distance: float = 3.5, vacuum: float = 15,
pbc: list = [True, True, False], reorder: bool=True, shift_tolerance: float = 1e-5) -> Atoms:
"""
After searching the coincidenced supecell, we get the transition matrix(M for bottom, N for top), and make two supercells, and then try to match them.\n
Additionally, bottom, top, weight, distance, vacuum.\n
usage: updating...
Noted: the cell C = A + weight * [B - A], A - bottom, B - top
"""
# get the max, min z-position
bottom = bottom_sup.copy()
top = top_sup.copy()
min_z1, max_z1 = bottom.positions[:,2].min(), bottom.positions[:,2].max()
min_z2, max_z2 = top.positions[:,2].min(), top.positions[:,2].max()
bottom_thickness = max_z1 - min_z1
top_thickness = max_z2 - min_z2
#print(bottom_thickness)
#print(distance)
shift = bottom_thickness + distance
# add distance
bottom.positions[:,2] -= min_z1
bottom.positions[:,2] += shift_tolerance
top.positions[:,2] -= min_z2
top.positions[:,2] += shift
top.positions[:,2] += shift_tolerance
# generate lattice
new_lattice = zeros((3,3))
# C = A + weight * [B - A]
for i in range(2):
for j in range(2):
#print(i,j)
new_lattice[i, j] = bottom.cell[i, j] + weight * (top.cell[i, j] - bottom.cell[i, j])
new_lattice[2,2] = bottom_thickness + distance + top_thickness + vacuum
# combine the position and symbols information
all_positions = concatenate([bottom.positions, top.positions], axis=0)
all_symbols = bottom.get_chemical_symbols() + top.get_chemical_symbols()
# create new Atoms
stack = Atoms(symbols=all_symbols, positions=all_positions, cell=new_lattice, pbc=pbc)
stack.center()
if reorder:
stack = sort(stack)
return stack
print(build_supercells(bottom, top, j.M, j.N, j.angle, j._weight))
print(build_supercells(bottom, top, j.M, j.N, j.angle, 0.5))
print(build_supercells(bottom, top, j.M, j.N, j.angle, 0))
print(build_supercells(bottom, top, j.M, j.N, j.angle, 1))
Strain, Stress
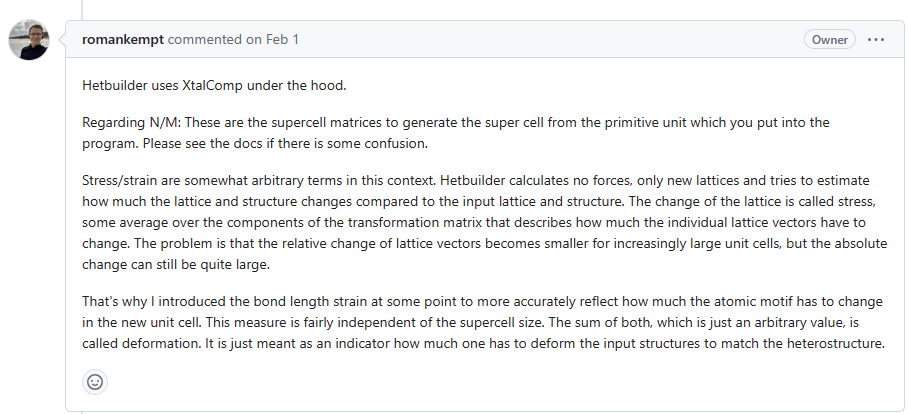
Some bugs
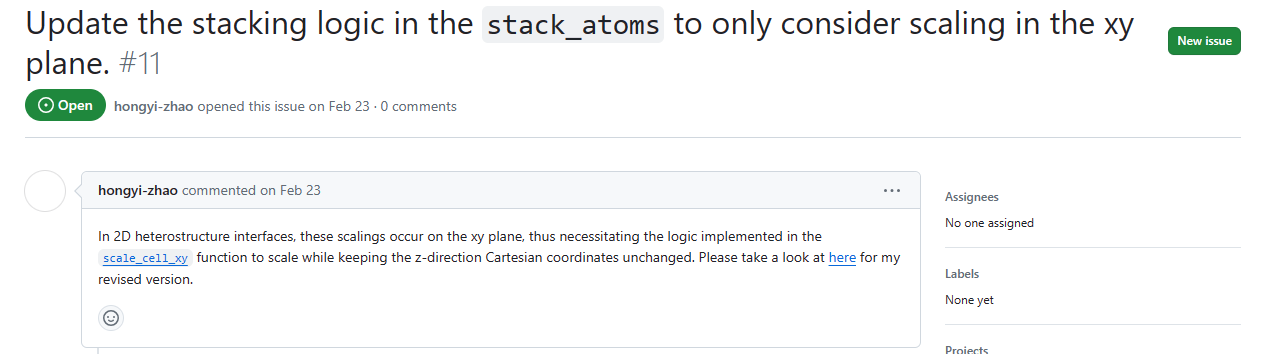
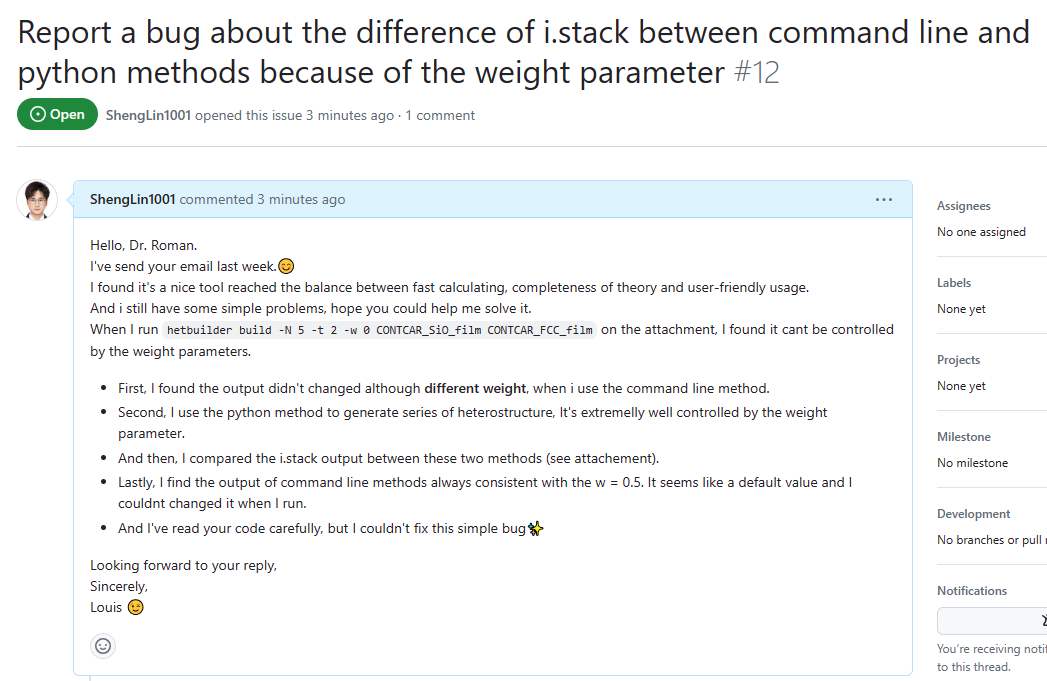

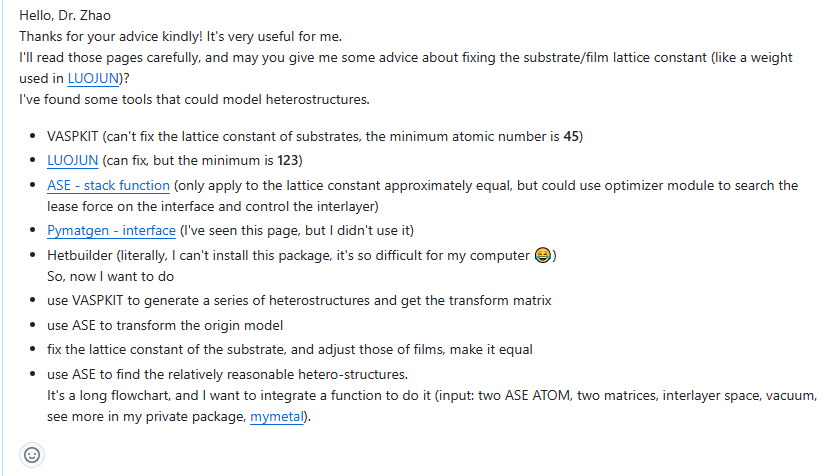
Fixed by change cli.py, it lose weight parameter when the run function are executed.
Next step
Zur algorithm
see Endnote/DFT-HCP-Au/Construct-Heterostructure/Lattice match: An application to heteroepitaxy
see Endnote/DFT-HCP-Au/Construct-Heterostructure/Computational Approach for Epitaxial Polymorph Stabilization through Substrate Selection
Interface-master
see Endnote/DFT-HCP-Au/Construct-Heterostructure/A brute-force code searching for cell of non-identical displacement for CSL grain boundaries and interfaces
InterMatch
see Endnote/DFT-HCP-Au/Construct-Heterostructure/High-throughput ab initio design of atomic interfaces using InterMatch
Coincidence-lattice
see Endnote/DFT-HCP-Au/Construct-Heterostructure/Coincidence Lattices of 2D Crystals: Heterostructure Predictions and Applications
Further thinking
Het2D maybe useful!
Please indicate the source when reprinting. Please verify the citation sources in the article and point out any errors or unclear expressions.